Jury Statement
The “SCREEN SCRYING” project plays with a surprising combination of complex scientific analysis with largely artistic visualization methods. The overall effect is an emotionally charged constellation of images which ask questions of both rational and irrational methods of analysis. This look at modern aesthetics of chaos and order take place in the highly charged context of the global threat posed by a dangerous pathogen that brings with it individual and societal upheavals—up to and including existential risk.
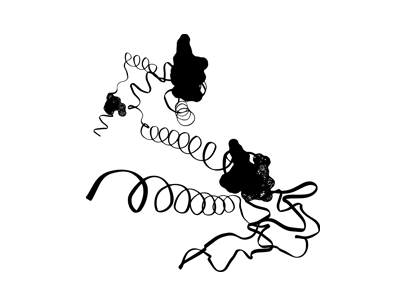
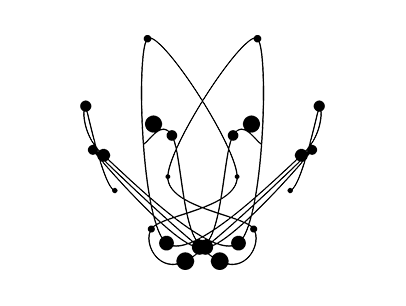
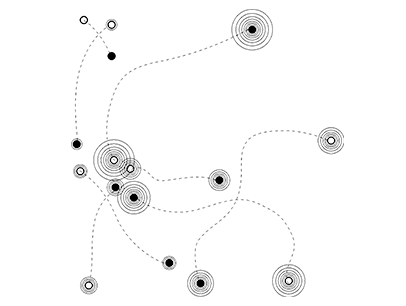
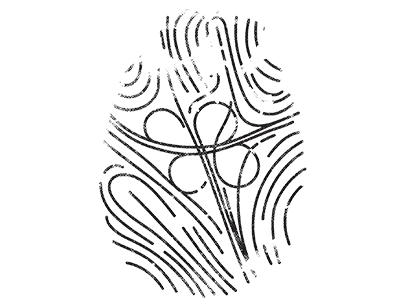
Ali Razzak, as a studied bioinformatics scientist, has not only mastered the vocabulary and the methods of the scientific analysis of dangerous pathogens, but also deals with the effect and meaningfulness of these methods as well as their visual forms. He demonstrates an understanding of pathogen representation and, above all, their effects in a social and historical context. In a quasi-Dadaist repositioning of the genetic codes of pathogens, by using several precisely defined steps and assigning meaning to randomly found letter sequences, his artwork generates the effect of looking into a crystal ball.
These 45 symbols are not intended to scientifically represent or otherwise accurately depict the structure of pathogens, but rather they reflect the current methods and tools available to us as a society to somehow imagine or divine the future during such an uncertain time. Above all, they reflect our fears and hysteria towards the invisible and omnipresent threat. The multi-layered and in-depth complexity of this concept, the contrast of logic and fantasy combined with the high visual quality of the 45 images impressively illustrate what Albert Camus, philosopher of the absurd, called the “clash of the irrational with the violent desire for clarity that is loud in the deepest inner human being”.
Artist Statement
Despite innumerable scientific advances, the arms race between human and pathogen is as competitive as ever, highlighting the sophisticated processes that govern microorganism behavior. To seek an understanding of the networks that animate pathogens, scientists have erected innumerable databases of molecular information. This has attracted the employment of computational tools to glean insight from that data, conjuring landscapes of projection maps. However, the intelligence that underlies these machine algorithms is often nebulous and can sometimes obfuscate meaning instead of elucidating it.
In recent times, international affairs were thrown into chaos and global operations were halted in the face of COVID-19. While times of crisis cast fear into an uncertain future, they also served as a reminder of monumental pandemics of the past. Historically, pathogens have devastated entire civilizations and pushed humans to the brink of extinction. The spread of COVID-19 invites new examination of pathogenic catastrophes as scientists take stock of what information, methods, and technology exist to address the crisis.
Extrapolating data about COVID-19 often calls for the implementation of tools which manifest a logic unto themselves. For example, what if the 45 worst human pathogens were coded through the use of sentiment analysis? We could change the individual makeup of each molecular depiction, give them names, and affect their depictions by assigning them positive/negative emotions. So often, scientific languages unravel cohesion in the attempt to create it. This project seeks to demonstrate how data-driven examinations of biomolecular catastrophes can recapitulate hysteria instead of resolving it.
Process
1. The first 25 protein molecule sequences[1] of human history’s 45 deadliest microorganisms (e.g. Serratia spp.), were retrieved from a national biomolecular database[2] .
2. Each molecular sequence was catalogued.
3. From that catalogue, a program which evaluates and assigns “sentiment analysis [to] single words”[3] was used to assign a positive (happy) or negative (sad) association to different pieces of the molecular structure’s code. (For example, in the sample data set Serratia ssp., “…WGARTLB…”, the word “ART” was identified by the program as “positive”.)
4. The ribbons and folds, etc. of a protein all have meaning in molecular biology, but the following process involved taking a construct that meant something in science and using it as a backbone to represent something in art. The representative molecule model[4] for each pathogen was then visualized through a software program.[5]
5. The words found in each molecular sequence were rendered as “wireframe” if positive or “opaque” if negative, and rendered to various “resolutions” depending on the degree of negativity or positivity. Thus, new images of the protein molecules were formed through a language of computer connotation.
_______
[1] .fasta files
[2] rcsb.org
[3] data.world/crowdflower/sentiment-analysis-single-word
[4] (e.g. 6RAO.pdb file)
[5] Visual Molecular Dynamics (VMD)
Ali Razzak
Ali Razzak is a structural bioinformatician and data scientist who was born in Iraq, raised in New Zealand, was a PhD candidate in Switzerland, and now resides in the United Kingdom. Outside of science, his interests lie in graphic and web design. He utilizes digital mediums to traverse migration, data mining, and fashion culture discourse. He is passionate about using unconventional tools to produce transdisciplinary discourse.